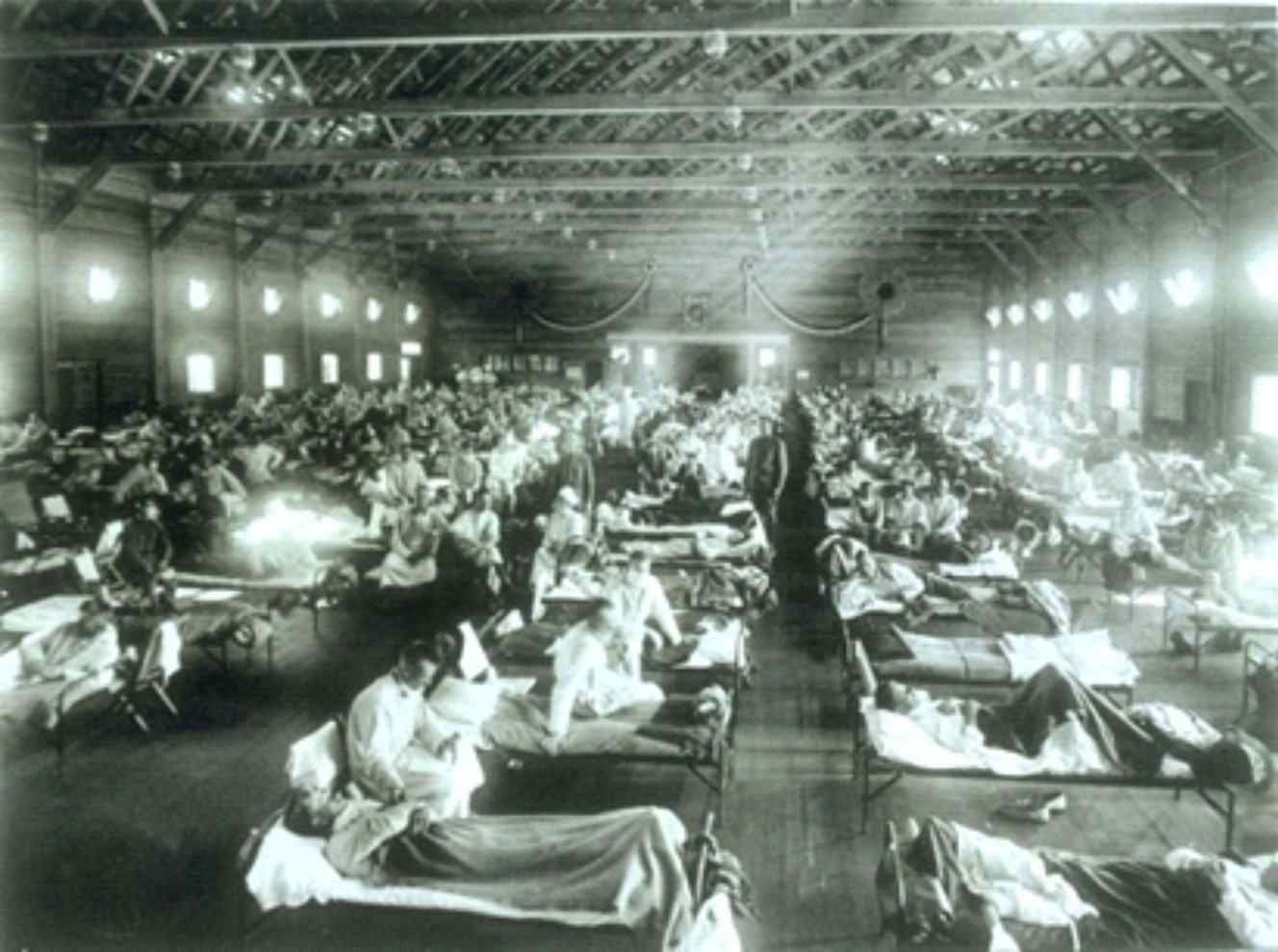
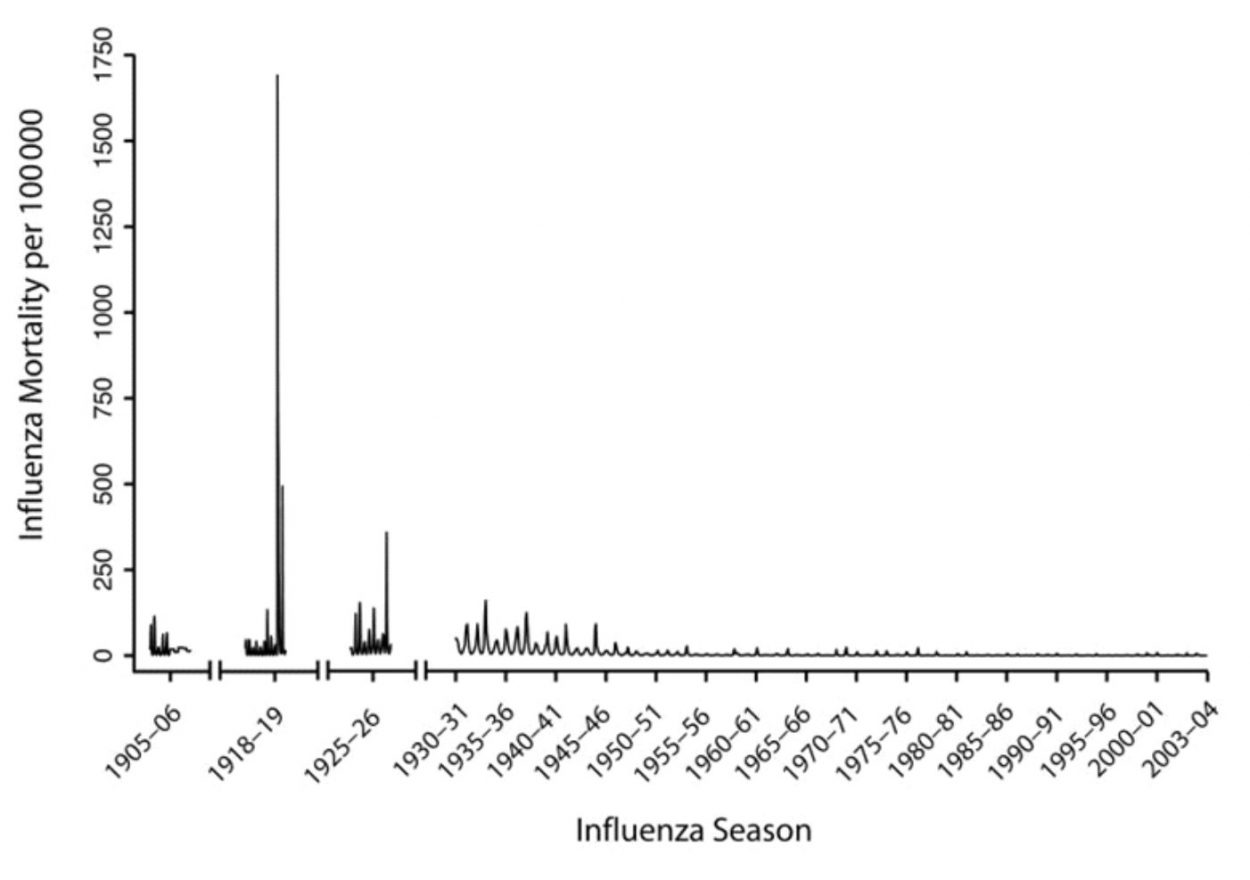
This year’s influenza season is on track to be the worst since the 2009 swine flu pandemic, so far claiming 114 pediatric lives. As tragic as this has been, it needs to be put in perspective. Let’s not forget the Spanish Flu Pandemic of 1918 that killed an estimated 500,000 Americans in just 18 months, with a worldwide death toll of 20 to 50 million.
Now, 100 years later, the world’s population has quadrupled, and global travel is much more common – two conditions that favor the spread of diseases. Using simple math and keeping all other factors equal, a similar pandemic could cause 80 to 200 million people to perish (one out of every 650 Americans). Some may naively counter that advances in medicine would prevent such a catastrophe, but our current influenza vaccines, therapeutics and diagnostics will likely not work well enough against a new strain of influenza virus to be very impactful.
Today’s vaccines target only circulating strains, and since the 2003-2004 flu season, the overall adjusted vaccine effectiveness has ranged from 10-60 percent, providing a modest to decent level of protection. However, the yearly vaccine is not expected to protect us against a novel influenza virus. From a therapeutic perspective, the influenza drugs Tamiflu and Relenza only slightly lessen the severity of the disease and curtail the duration of symptoms by about one day when administered within 48 hours of symptoms onset.
Although these drugs have been shown to be effective on the currently circulating strains, there is no guarantee that they will work on the next pandemic strain. Further, our point-of-care diagnostics that are easy-to-use are all “closed-access” systems, which means that the manufacturers have pre-configured their tests to only detect currently circulating strains.
These technologies do not have the flexibility to be quickly modified to detect a novel strain. If these tests return a negative result, clinicians will be forced to send collected samples out for testing that can be customized using the expertise of a specialist. Such tests generally have a turn-around time of at least 24 hours – far too long for informing decisions in an emergency room setting.
In the event of a deadly outbreak, emergency rooms would be overwhelmed, and physicians would not be able to easily discriminate between the individuals who are in the early stages of being infected with the pandemic strain versus those who are infected with seasonal influenza, as the symptoms will likely be nearly identical. Once an ER becomes overcrowded, the pandemic virus will have an easier time infecting its next victims, and our emergency rooms will turn into depots for rapid viral spread.
Despite 100 years of preparation since the deadliest pandemic in humankind’s history, we still don’t have the tools to significantly reduce the impact of the next deadly influenza pandemic. Anthony Fauci, director of the NIH’s National Institute of Allergy and Infectious Disease; Anne Schuchat, director of the CDC and Bill Gates have admirably been ringing this alarm bell for years. On the private side, The Coalition for Epidemic Preparedness Innovations (CEPI) – founded by the Bill & Melinda Gates Foundation, governments of India and Norway, medical research charity Wellcome and World Economic Forum – has $460 million in dedicated funding from private, public and philanthropic organizations.
These monies are being directed toward research to develop faster ways to make vaccines, because the current strategies are too slow. On the public side, this year’s influenza season has spurred some U.S. senators to propose legislation that would provide $1 billion over five years to the NIH for the development of a universal vaccine against influenza, which would be a significant increase over the $64 million that was awarded to researchers in 2017. Whether Congress approves such spending is yet to be determined, and these goals of developing vaccines that are more quickly manufactured or a universal vaccine / drug are extremely lofty. Even if millions to billions are injected for research, there is no guarantee of success. In fact, the chances of success are actually quite small, but this doesn’t mean that the pursuit is not worthy.
With all of the past support for better diagnostics, never has a government funding agency (e.g. NIH, NSF, BARDA, DARPA) or a private philanthropic group like CEPI released a significant funding announcement that requests the development of “open-access” diagnostics. It is time that this changed. Our best chance to successfully minimize the impact of the next pandemic is to start immediately funding the development of open-access diagnostics that are fully automated and could be placed at the point-of-care (POC).
Although the world is in desperate need of easy-to-use, open-access diagnostics, none have been commercialized yet. With that said, one is under development, and possibly other companies are in pursuit of the same goal. To be successful, the developed system needs to be designed largely foolproof so that the standard test for seasonal influenza (or other targeted pathogens) is not impacted if “pandemic mode” of the open-access system is activated.
Activating pandemic mode involves an end-user adding newly synthesized pandemics-specific reagents onto the instrument such that it can access these reagents to broaden its screening capability. The step of adding these reagents must be simple – ideally done in less than two minutes – by a single person who aseptically provides sufficient liquid reagents into an existing empty reservoir(s) for the automated instrument to access.
Ideally, this reservoir(s) is large enough to hold sufficient reagents so that the instrument can process hundreds – if not thousands – of samples before reagent replenishment becomes necessary. Once the reagent is added, other users of the instrument would not need to change how they interface with the instrument, aside from selecting “pandemic mode” on the instrument’s touch screen monitor to instruct it to use these newly added reagents for expanded screening.
To choose the base chemistry to be used in an open-access diagnostic instrument, it needs to be the TaqMan® PCR chemistry for numerous reasons. First, it is considered a gold standard chemistry and has been used in molecular diagnostics for over a decade. Second, there are numerous TaqMan reagent manufacturers, including Integrated DNA Technologies (IDT), LGC Biosearch Technologies, Thermo Fisher Scientific, Sigma-Aldrich, Bio-Rad Laboratories, Eurofin Genomics and Operon Technologies to name a few.
Any of these companies could provide pathogen-specific reagents for an open-access instrument that utilizes TaqMan chemistry. It is critical to understand this point, as open-access allows third parties to manufacture reagents for an instrument that’s made by a different company. This effectively reduces the bottleneck of relying on a single manufacturer to provide reagents to institutions in the event of a deadly virus emerging.
TaqMan chemistry is also low cost and can return results in about an hour, which is fast enough to inform quarantine decisions in an emergency room. TaqMan assays can be synthesized very quickly, and two of the top manufacturers could make sufficient reagents (e.g. 500 uL of a 10X assay) for every major hospital and medical clinic in the United States in three days. Lastly, these reagents can be easily shipped overnight at ambient temperature in a padded envelope to help achieve the goal of getting pandemic detection capabilities in a POC setting within two weeks of sequencing the newly emergent causative pathogen.
For open-access technology to be the most impactful, every hospital and sizable medical clinic needs to have an instrument in place, being used on a daily basis to detect common pathogens (not necessarily influenza). In the event of an outbreak, the CDC or an academic lab would determine the sequence of the new pathogen’s genetic code within one to two days. Bio-informaticists could use this information to design TaqMan assays that are specific for the pathogen’s genetic code in just a few hours.
The CDC would effectively bless these designs (and the FDA would provide emergency use authorization), and reagent manufacturers could begin synthesizing new TaqMan assays that are specific to the new pathogen. Hospitals that are running open-access instruments would order reagents from one of the many TaqMan reagent manufacturers, and they would likely receive these reagents within 48 hours and be successfully running pandemic-specific tests just minutes after delivery.
It is doubtful that any strategy can completely avert a pandemic since, by nature, these are generally worldwide events. However, now more than ever, the government needs to have the foresight to recognize the need for open-access technologies and provide a funding environment that encourages innovation in this area. We need to act now so that new technologies can be developed, penetrate the market and begin being used on a daily basis for the diagnosis of everyday pathogens. These instruments must be ready to go to work when the next new threats to humanity arise.